Robust Geometric Graph Kernels for Bioimaging Applications

Project responsible: Aasa Feragen
Postdoctoral Fellow, Machine Learning and Computational Biology Research Group, Max Planck Institute for Intelligent Systems, Tübingen.
Freja Fellow and Assistant Professor, DIKU, University of Copenhagen
Email: aasa.feragen@tuebingen.mpg.de
Mail address:
MPI for Intelligent Systems
AG K. Borgwardt
Spemannstr. 38
72076 Tübingen
Germany
Can we save lives with mathematics and computer science?
Yes, we can!
|
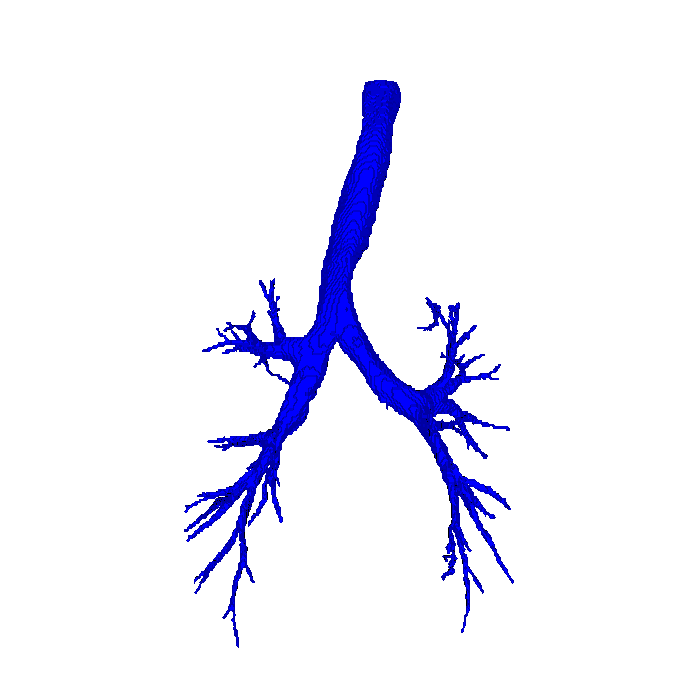
The transport systems in your body form geometric networks, which transport, for instance, blood, air or nerve impulses. The functional capacity of such a network is often closely connected with the functional capacity of the organ it is part of: If the airways in your lung do not transport air, then your lung cannot breathe. If the connective network in your brain is damaged, you lose cognitive function.
|
|
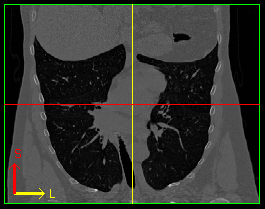
We can study anatomical networks through medical images, where information about blood vessels, airways or brain connectivity is extracted from scans. In this project, I will study how these networks differ in order to classify medical images of lungs and brains with respect to various illnesses, such as smoker's lung (Chronic Obstructive Pulmonary Disease), stress and multiple sclerosis.
|
|

|
In order to learn something from a large set of images, networks or, say, airway trees, we need to be able to compare them. One way of doing this is by imagining a large "space of airway trees". The points in this space are now airway trees, and the idea is that airway trees which are "close together" in the airway tree-space should also be "similar" in other ways. Often, "similar" is a measure of disease that is hard or expensive to determine, and the goal of research is to find a good measure of "close together" which is easy and cheap to compute while capturing the essence of the disease similarity.
|
|
A kernel is a particular example of such a measure of whether two airway trees or brain graphs are "close together", and the main goal of this project is to develop a framework that will allow us to use kernels for very different anatomical networks in a fast and cost-effective way. Once we have such a kernel, we automatically gain access to machine learning algorithms for classifying new airway trees as more similar to healthy or ill airway trees, or for discovering new subtypes of illness. In this way, mathematics and computer science can actually, in the final end, be part of developing more efficient diagnostic technology which saves lives. Isn't that cool?
|

|
Planned activities
May 12-14: Speaker at the mini-symposium Detection and Analysis of Blood Vessels and Tree Shapes at the SIAM Conference of Imaging Science, Hong Kong
February 10-14 2014: Speaker at the MBI Workshop 2: Morphogenesis, Regeneration, and the Analysis of Shape
December 18: Weigelworld talk.
December 5-11: Presenting the paper Scalable kernels for graphs with continuous attributes at NIPS 2013.
More information
Detailed project description
Project publications
Activities
Collaboration Partners
The Machine Learning and Computational Biology Research Group, MPI Tübingen.
The Image Group, Department of Computer Science, University of Copenhagen
Acknowledgement
This project is funded by The Danish Council for Independent Research | Technology and Production Sciences.
|




